
Validation of liquid chromatography mass spectrometry (LC-MS) methods
3.2. Experiment setup and evaluation of the data
Experiment planning for evaluation of
http://www.uttv.ee/naita?id=23307
https://www.youtube.com/watch?v=PdLsxDExgV0
1) Type of calibration samples
When choosing the proper quantitation method, we can choose between calibration samples (calibration standards, calibrants) containing matrix and calibration samples that are matrix free. In case of LC-MS analysis, we should prefer samples containing matrix, in order to take into account possible matrix influence on the ionization of the analyte. Blank matrix extracts of as similar as possible matrix type as the sample are suitable for this.
If the sample to be analysed is diluted prior to the analysis, the matrix concentration in the matrix-matched calibration standards should be diluted proportionately so that the matrix amount in each analyzed sample is constant.
If calibration solutions in solvent are used, a comparison of calibration graphs in matrix and in solvent should be carried out. (1)
2) Concentrations of calibration samples
The highest and lowest concentrations of the calibration samples should be appropriate for the method, keeping in mind the predicted variation of the analyte concentration levels in the samples. As a minimum, 6 different concentrations are necessary according to most validation guidelines. This is also acceptable for statistical tests carried out later for linearity evaluation, where 4 degrees of freedom is considered minimal. However, as we do not know the span of the at this step of the validation, some concentrations might fall out of the linear range. Therefore, using 10 concentration levels encompassing the expected linear range is recommended. Moreover, concentrations should be approximately evenly spaced over the chosen concentration range, to ensure that the different parts of a calibration graph are covered with approximately the same density of data points.
3) Measurement protocol
For LC-MS, the order of measuring the solutions in the series and the number of replicate measurements with every solution, is important due to the possible drift or contamination of the instrument. Consequently, the analysis order of calibration samples should be random.
It is useful to analyze calibration solutions in a manner as similar as possible to the unknown samples, i.e. calibration samples should be in random order and placed between unknown samples in the . Calibration samples should be analyzed at least twice (and average values should be used in linearity calculations).
In the following video, the preparation of matrix matched calibration samples on an example of pesticide analysis in tomato is shown.
Carrying out the experiment for linearity evaluation
http://www.uttv.ee/naita?id=23480
https://www.youtube.com/watch?v=x8KaQ7aC_mI
Evaluation of linearity from the experimental data
For quantitative analysis, the calibration data is plotted on a calibration graph, where the concentrations are on the x-axis and the signals are on the y-axis. (2)
From this plotted graph we can have the first evaluation of the linearity by using the visual evaluation approach. In addition, calculation of the residuals can also be useful, due to the fact that the residuals show a more clear picture if the applied linear model actually fits the data.
Evaluation of linearity
http://www.uttv.ee/naita?id=23351
https://www.youtube.com/watch?v=4c3EMRpFDf0&t=29s
Absolute residuals are found as the difference between the experimental (yi) and calculated (ŷi) signal values: yi– ŷi. In addition, relative residuals can be used (Eq 1).
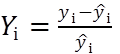 (Eq 1)
(Eq 1)
For more complex cases, where a linearity cannot be confirmed by neither a visual evaluation nor residuals, statistical approaches can be of help.
Statistical approaches for evaluation of linearity *Note 1
http://www.uttv.ee/naita?id=23683
https://www.youtube.com/watch?v=QFNuo-Jk2Ws
*Note 1: Please see comments in the Mandel’s test section below on the simplification in the video. The Lack-of-fit equation in this video should be understood the same way as (Eq 2) in the Lack-of-fit section below.
Different expressions of signal values are used for statistical approaches:
yij is the experimental signal value at the concentration level i for replicate measurement j,
ȳi is the average value of the experimental signals from p replicate measurements at the concentration level i,
ŷi is the signal value at the concentration level i, calculated using the calibration function.
In addition, n is the number of concentration levels and p is the number of replicate measurements at each concentration level.
Tabulated F-values can be found here: http://www.itl.nist.gov/div898/handbook/eda/section3/eda3673.htm
The Ftabulated can also be found in excel using the following function:
=F.INV.RT(α;DoF1;DoF2),
where:
α is the probability of the F distribution (on 95% confidence interval α=0.05*),
DoF1 is the number of degrees of freedom of the numerator MSSLoF (n-2),
DoF2 is the number of degrees of freedom of the denominator MSSerror (n*(p-1)).
As a generalisation, the number of degrees of freedom is equal to the number of data points minus the number of parameters calculated from the data. Example: In simple linear regression of the type y = b0 + b1 · x DoF is typically n – 2, where n is the number of data points. The „2“ means that two parameters are found: slope and intercept. If the intercept is forced to zero, i.e. if the regression has the form y = b1 · x then DoF = n – 1, because only one parameter is found.
*The 0.05 is found from 95% as follows: (100% – 95%) / 100% = 0.05.
Lack-of-fit test
IUPAC validation guideline suggests using the lack-of-fit test. The extent of deviation of the points from the line caused by the random scatter of the points is estimated from the replicate measurements (mean sum of squares of random error (MSSerror)).
This is compared to the extent of deviation of the points from the line caused by the mismatch of the calibration model (mean sum of squares due to lack of fit MSSLOF).
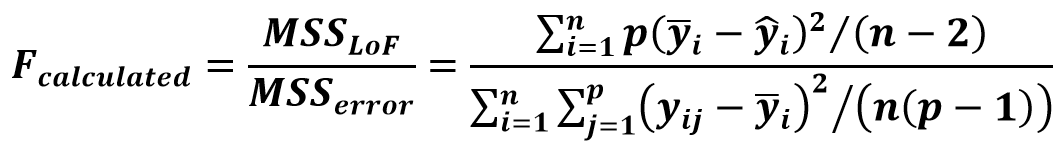 (Eq 2)
(Eq 2)
If the Fcalculated is higher than the Ftabulated, the model cannot be considered fit for the data, because the unexplained variance in the model is too big.
Goodness-of-fit test
The goodness-of-fit test uses the mean sum of squares of the factors (MSSfactor) describing the variance described by the model and the mean sum of squares of the residuals (MSSresiduals).
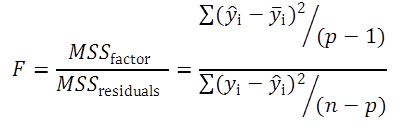 (Eq 3)
(Eq 3)
If the Fcalculated is higher than the Ftabulated, the model differs systematically from the data.
Mandel’s fitting test
This test compares the fit of two models: the fit of a linear model (Sy1) with the fit of a nonlinear model (Sy2). In this case a three-parameter model, parabola, is used. Three-parameter model is suitable for almost all cases. For both of the models, the residual standard deviation is found using this equation:
 (Eq 4)
(Eq 4)
, where n is the number of calibration points (assuming one point per calibration level, i.e. p = 1). In case p > 1, it is assumed that all concentration levels have the same number of replicates. In many materials n·p is denoted by N.
The Fcalculated is found:
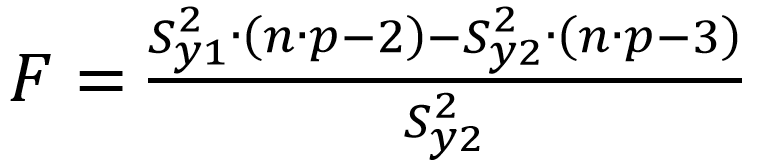 (Eq 5)
(Eq 5)
If the Fcalculated is higher than the Ftabulated, the linear model cannot be applied.
Please note that in different materials the Mandel’s fitting test is presented at different level of rigor. In the video, a simplified version is presented, assuming that p is always 1 and disregarding the difference of degrees of freedom between linear and nonlinear model, using n – 2 in both cases. A more rigorous version, presented here. In practical application, the differences between the two are not big.
Intercept
One important issue concerning the calibration graph is how to handle the intercept. As usually the linear calibration graph model is used, the model has two parameters: a slope and an intercept. A slope gives us the estimation of the of our method (signal per one concentration unit, see section 3.4), while an intercept shows the estimate of the signal for a blank sample.
For most of the HPLC detectors, including the MS, it is fair to assume that the sample without an analyte gives no signal. Also for most detectors, intercept is not consistent with the physical model behind the calibration principle. For example in the case of HPLC-UV/Vis, the calibration model should follow the Beer´s law:
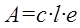 (Eq 6)
(Eq 6)
where A is the measured absorbance, c is an analyte concentration, l is the optical path length and e is the molar absorption coefficient. Therefore, as the physics behind the HPLC-UV/Vis signal does not contain an intercept, it is worth checking if the intercept is statistically significant at all.
The statistical way to evaluate the importance of an intercept would be via a t-test. In order to carry out a t-test, the linear regression is with an intercept, i.e. in the form LINEST(Y1:Y2; X1:X2; 1; 1), and the obtained intercept value is compared to zero taking into account the standard deviation of the intercept and the number of points on the calibration graph. However, a simpler method can be used that is based on the assumption of normal distribution and also assuming that there is a sufficient number of points on the calibration graph. In this case, the t-value is substituted with 2 (referring to the 95% confidence level in normal distribution).
If
Intercept < 2 · Stdev_intercept
then it can be assumed with 95% confidence that the intercept is insignificant and can be disregarded in the calibration model. The following form of the LINEST spreadsheet function is used in this case: LINEST(Y1:Y2; X1:X2; 0; 1). Setting the third parameter in the function to zero forces the intercept to zero.
If an intercept, however, is statistically significant, but a physical model behind the detection does not contain an intercept, it may be an indication of problems. Most commonly:
- a linear model is fitted to data that are in fact nonlinear (e.g. saturation of signal at higher concentrations);
- blank samples produce signal because of a carryover, contamination, etc.
Both of these should be carefully studied and if possible, removed.
Evaluation of linearity (visual evaluation, residuals)
http://www.uttv.ee/naita?id=24974
https://youtu.be/A7hsHZXMsbY?si=Qikjm-e_LoMpDbDg
Evaluation of linearity (lack-of-fit test)
http://www.uttv.ee/naita?id=32138
https://youtu.be/l07_KulyYoc
***
(1) Both standard (in solvent) and matrix—matched calibration curves should be constructed. If the matrix does not interfere with the analysis and the use of a standard (in solution) calibration curve is justified, the slopes of these two graphs should not differ statistically. This can be shown using a t-test. In order to do so, the residual variances of the two graphs should be equal. This can be confirmed by using an F-test.
An example where the slopes of the two graphs do not differ statistically and the use of a standard calibration graph is justified can be found in .
An example where the matrix interferes with the analysis and a matrix matched calibration is used can be found in .
(2) Calibration curve definition by VIM – the expression of the relation between indication and corresponding measured quantity value. “The term “curve” implies that the line is not straight. However, the best (parts of) calibration lines are linear and, therefore, the general term “graph” is preferred.” [http://www.fao.org/docrep/w7295e/w7295e09.htm#TopOfPage]


