Mutations in ribosomal protein uS5 alter translation fidelity and mutagenesis in Pseudomonas putida
Jürgenstein K, Ilves H, Luhaäär C, Brauer A, Remme J, Kivisaar M.
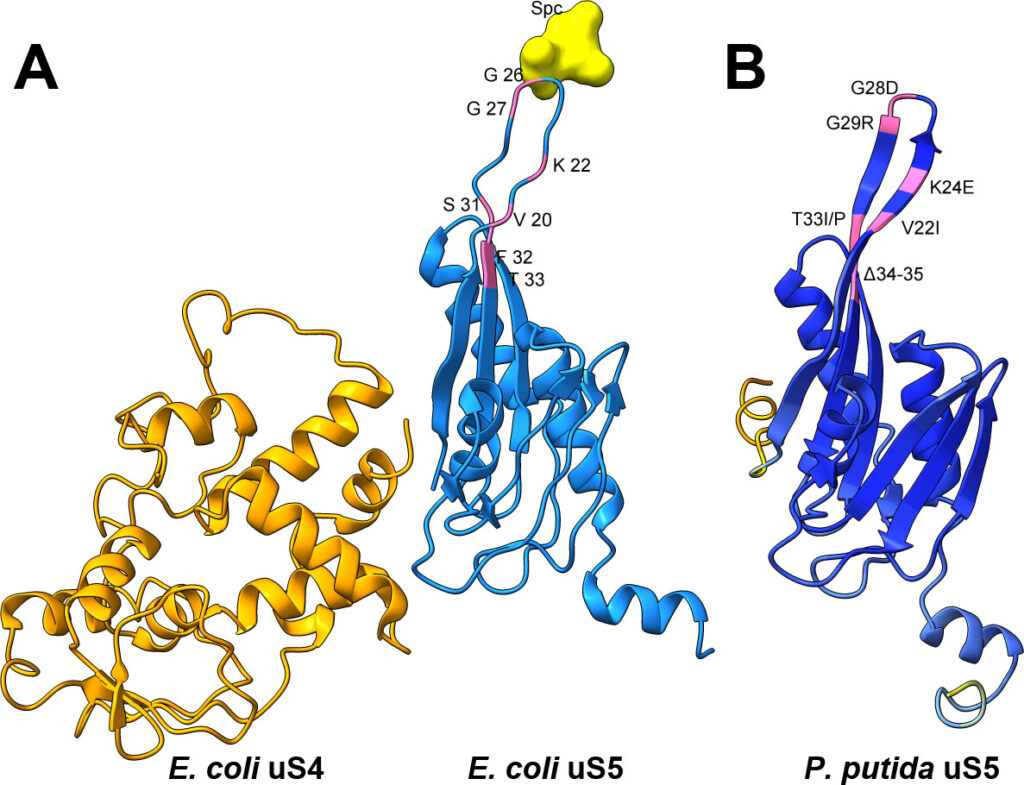
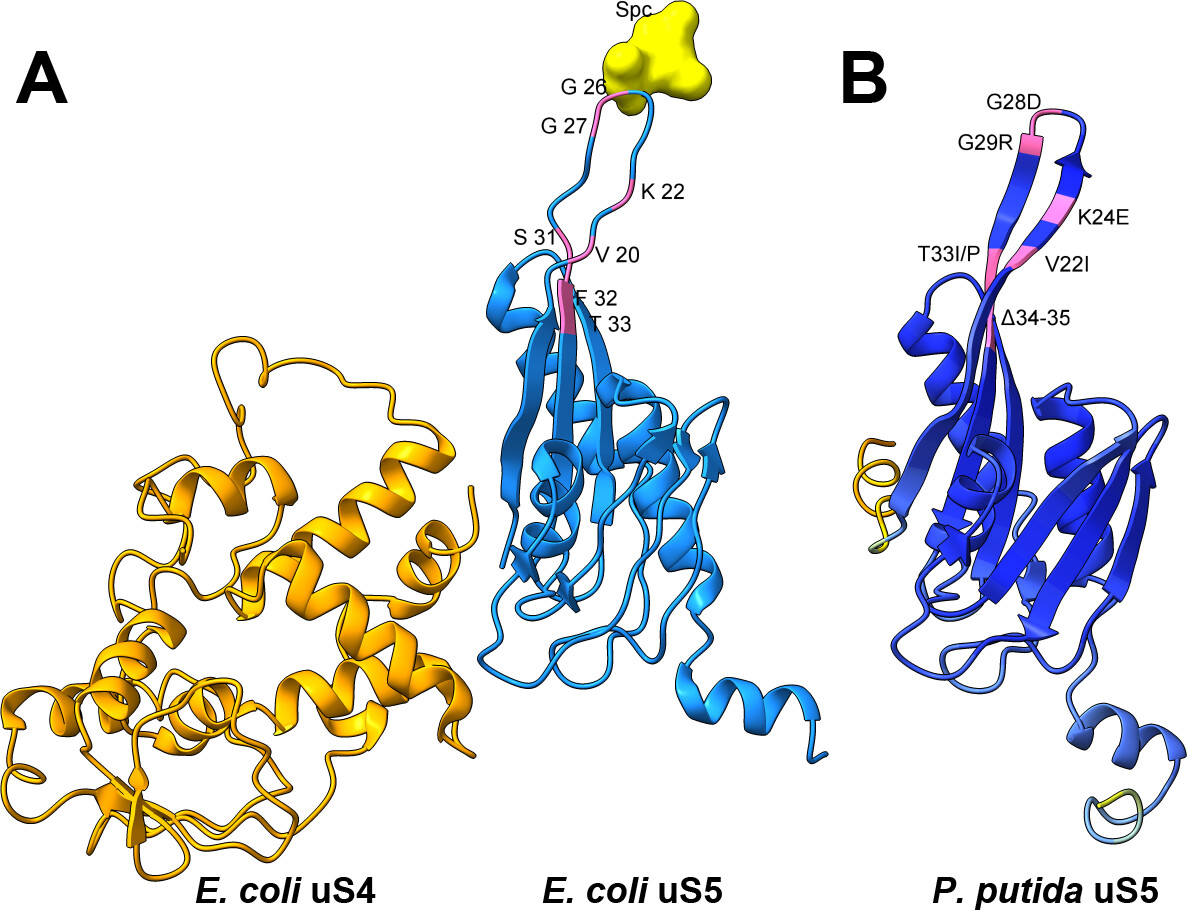
Errors in protein synthesis can influence cellular fitness and, in some cases, affect genome stability. While this connection has been explored, most investigations have focused on Enterobacteriaceae such as Escherichia coli, leaving open the question of how translational fidelity shapes mutagenesis in other bacterial groups. Would a metabolically versatile, stress-tolerant soil bacterium behave differently? We focused on the small-subunit ribosomal protein uS5 (rpsE) because, despite its role in decoding accuracy, its potential as a mutator locus has not been evaluated. To explore this, we isolated several mutants of ribosomal protein uS5 (rpsE) in Pseudomonas putida, all carrying amino acid substitutions or deletions in loop 2, via spectinomycin selection, and then quantified their effects on frameshifting and stop-codon readthrough using dual-luciferase reporters. In parallel, we measured the rate and spectrum of spontaneous rifampicin resistance (rpoB) mutations. The fitness costs of these uS5 alleles were also assessed through growth and stress tolerance assays. Several mutants displayed divergent decoding phenotypes, including both error-prone and error-restrictive profiles, and the increase in translational errors was often, but not universally, coupled with elevated mutation rate. This approach probes which uS5 perturbations confer a bona fide mutator phenotype and how decoding errors bias mutation outcomes. By extending fidelity mutagenesis studies beyond enterobacteria, our work identifies uS5 as a previously unrecognized mutator locus in P. putida and illuminates the nuanced coupling of translation accuracy and evolvability in a key environmental microbe.
J Bacteriol. 2025 Nov 12:e0033425. doi: 10.1128/jb.00334-25