Hypoxia-induced ribosomal RNA modifications in the peptidyl-transferase center contribute to anaerobic growth of bacteria
Ishiguro K, Midorikawa K, Shigi N, Kimura S, Liiv A, Yokoyama T, Ito T, Shirouzu M, Remme J, Miyauchi K, Suzuki T.
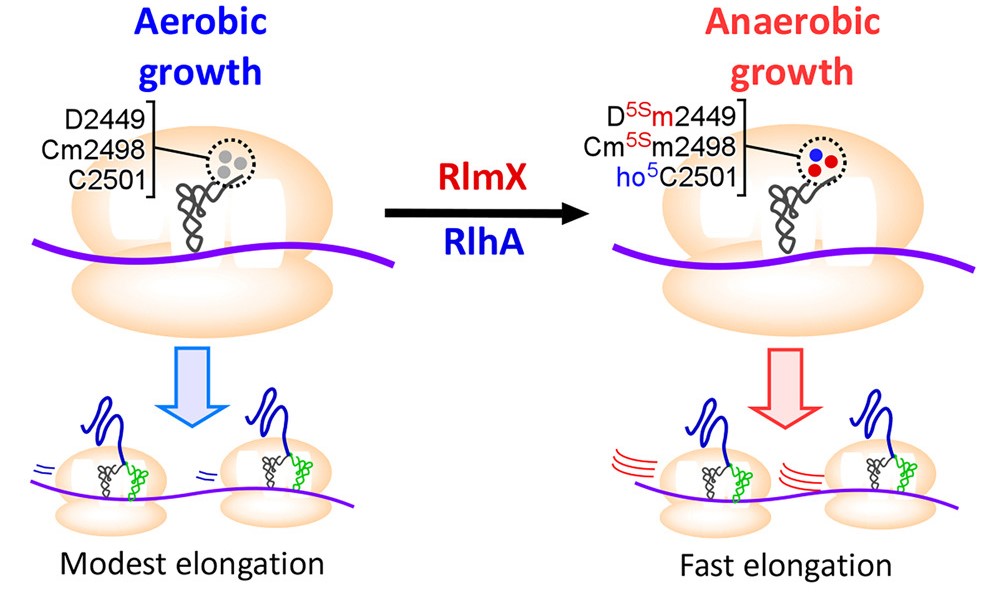
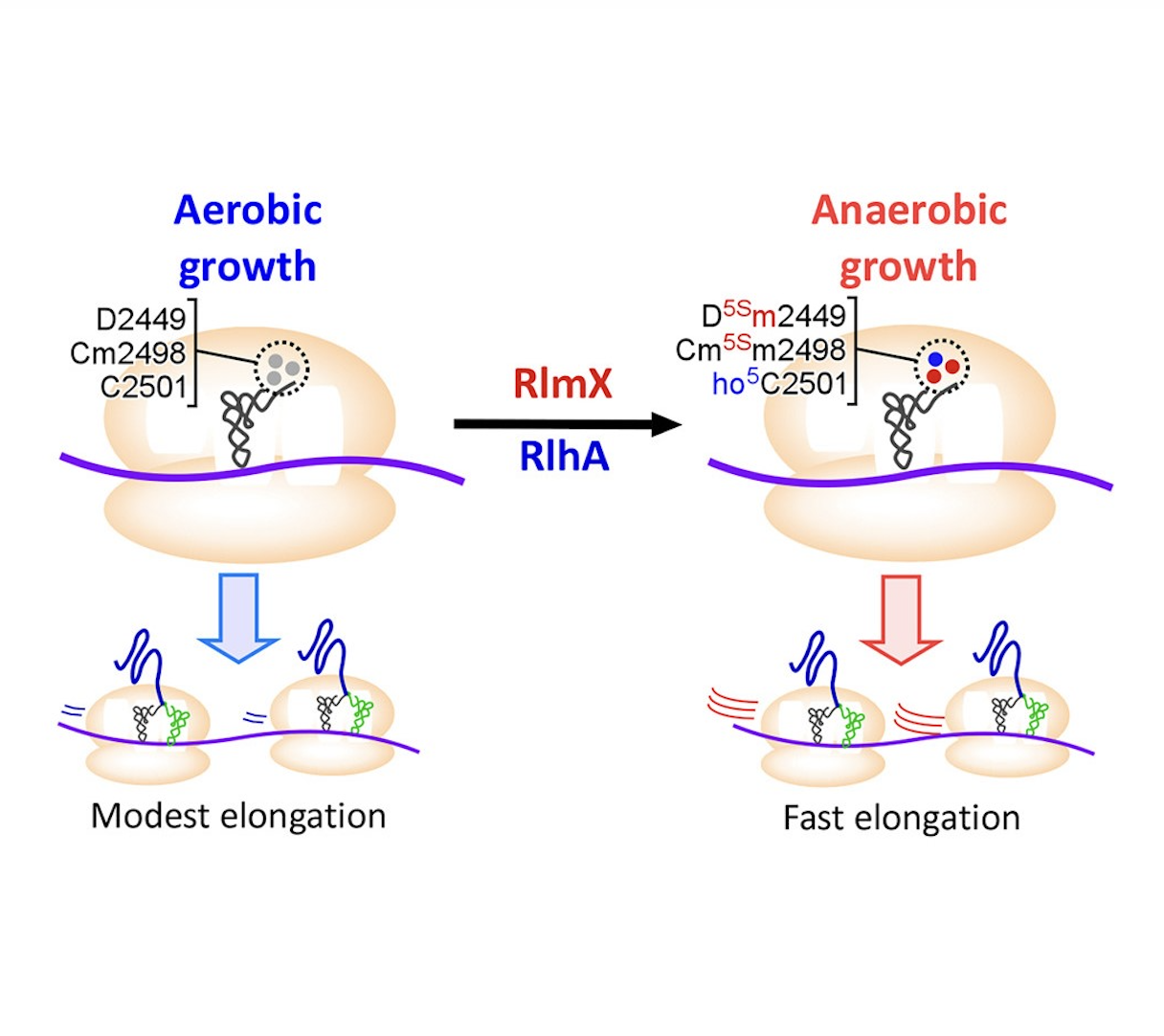
Ribosomal RNAs (rRNAs) contain various modifications that play critical roles in ribosome assembly and function. Here, we discovered two stereoselective methylations of the rRNA backbone in the peptidyl-transferase center (PTC) of the 50S subunit of Escherichia coli cultured under anaerobic conditions. Methylation occurs at carbon 5′(S) of ribose moieties of dihydrouridine at position 2449 (D5Sm2449) and 2′-O-metylcytidine at position 2498 (Cm5Sm2498). We identified the rlmX gene, encoding a cobalamin-dependent radical S-adenosylmethionine (SAM) methyltransferase responsible for these methylations. Intriguingly, D5Sm2449, Cm5Sm2498, and 5-hydroxycytidine (ho5C2501) in the PTC were elevated under anaerobic growth conditions. A double knockout strain lacking rlmX and rlhA (responsible for ho5C2501) impaired anaerobic growth. Biochemical studies showed that these rRNA modifications stimulate protein synthesis. The cryoelectron microscopy (cryo-EM) structure of the ribosome indicated that these hypoxia-induced modifications stabilize the P-site and the PTC. These findings demonstrate that ribosomes are activated by hypoxia-induced modifications to enhance translational capability and thereby survival, under anaerobic conditions.