MRI bioinformaatika töögrupp
Research
1. Computational analysis of bacterial genomes and prediction of antimicrobial resistance.
We develop computational tools for automated analysis of bacterial genome sequences. This includes assembly, validation of species, check and elimination of contamination by other species, determination of MLST type, building a phylogenetic tree from core genome sequences, detecting the presence and intactness of known resistance genes, detecting and describing the context of resistance genes, detection and description of plasmid sequences. Some of the tools for this analysis have been developed earlier in our workgroup, e.g. StrainSeeker (Roosaare et al., 2017), PlasmidSeeker (Roosaare et al., 2018) and PhenotypeSeeker (Aun et al., 2018).
Large-scale analysis of bacterial genomes can be done with different purposes. The most common aim is the epidemiological analysis of the spread of bacterial strains or their resistance genes (Telling et al., 2018; Bilozor et al., 2019; Sepp et al., 2019; Telling et al., 2020, Aun et al., 2021).
Second frequent aim of bacterial genome analysis is a functional analysis of genomic variants that appear under strong natural selection in a certain environment. A good example of this kind of analysis is the paper by Jõers et al., 2019, where we describe which genes are mutated in a strain that is less responsive to muropeptides in the environment. Currently, our group is actively working towards developing statistical prediction models that would predict virulence, antimicrobial resistance or other properties of bacterial strains, based on their sequence.
2. Development of diagnostic tests based on DNA sequence.
For more than 10 years we have been studying how DNA molecules hybridize to their specific and non-specific targets. This is important for designing highly specific PCR primers or microarray probes. We have written several software packages facilitating genomic PCR primer design. Our workgroup has participated in the development of widely used Primer3 software and we host an official webpage for Primer3.
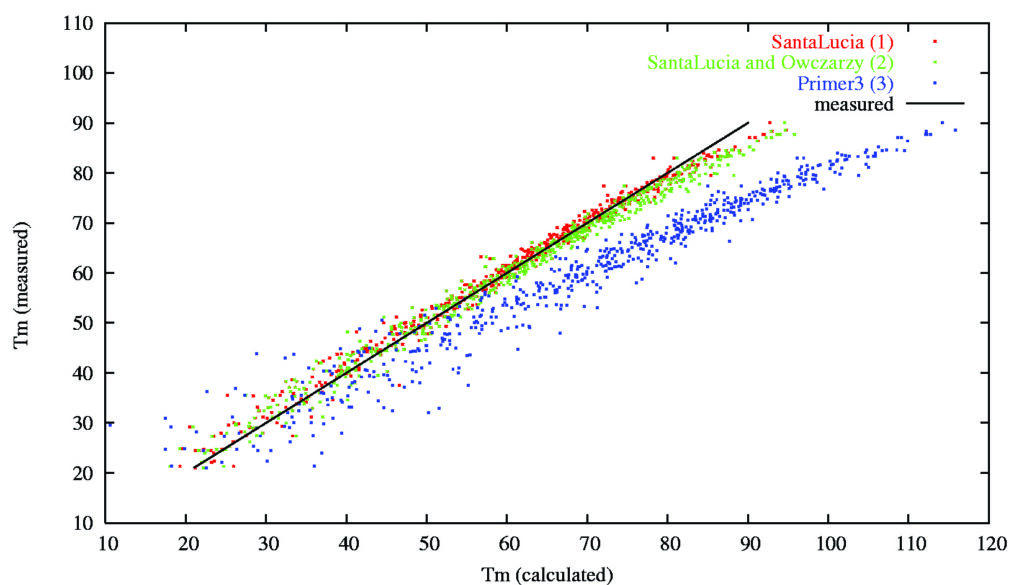
Prediction of primer melting temperatures (Tm) by old Primer3 (blue dots) and by modified Primer3 (green and red dots).
We have implemented our theoretical knowledge in practice in form of various DNA-based molecular tests. For example, we have developed diagnostic tests for respiratory disease pathogen detection for Estonian company Quattromed HTI (currently part of SynLab Eesti), blood sepsis-related pathogen tests for Dutch start-up company Microbiome Ltd (van den Brand et al., 2014) and food allergen detection test for Estonian biotech company Icosagen Ltd. We have developed a multiplex PCR tests that would enable faster detection of sequence types of food pathogen Listeria monocytogenes.
Currently the main focus is to develop novel diagnostic tests, based on next-generation sequencing data. A large advantage of DNA sequencing tests over PCR-based tests is that it can detect all pathogens simultaneously, unlike PCR that can detect only one pathogen per primer pair. We have developed a computational framework for a test that allows detection of sepsis pathogens directly from blood sample (using cell-free DNA).
Another area where WGS data can be used is food monitoring. Example applications are described in papers Raime and Remm, 2018 and Raime et al., 2020. In these papers, we described methods and proof-of-principle tests for the detection of potentially allergenic food components using food sequencing approach. We continue with development of methods allowing the detection of food pathogens, food origin and food composition.
3. Development of fast and original computational methods for the analysis of personal genomes.
Within the last ten years, we have developed several novel methods for the detection of variants from human personal genomes.
* FastGT (Pajuste et al., 2017) finds genotypes of all previously known SNV-s in a given personal genome in only 30 minutes.
* KATK finds all SNVs (known and novel) from personal genomes in 3 hours (Kaplinski et al., 2021).
* We can detect polymorphic Alu element insertion sites (Puurand et al., 2019),
* We can predict gene copy numbers, for example amylase gene copy numbers (Pajuste et al., 2023)
* We can predict copy numbers of variable number tandem repeats e.g. those in front of ACAN, MAO, TRIB3 genes (Örd et al., 2020).
* We can predict human chrY haplogroups from low-coverage sequencing data (Puurand et al., 2025)
Our algorithms use a k-mer based signature sequences which allows us to detect those variants in personal genomes ca 30 faster than traditional methods, without losing accuracy.